Antibody data
- Antibody Data
- Antigen structure
- References [4]
- Comments [0]
- Validations
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA017322 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA017322, RRID:AB_1848424
- Product name
- Anti-FAR1
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human FAR1, Gene description: fatty acyl CoA reductase 1, Alternative Gene Names: FLJ22728, MLSTD2, SDR10E1, Validated applications: WB, IHC, ICC, Uniprot ID: Q8WVX9, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.1 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references The origin of long-chain fatty acids required for de novo ether lipid/plasmalogen synthesis
The peroxisomal enzyme, FAR1, is induced during ER stress in an ATF6-dependent manner in cardiac myocytes
An autosomal dominant neurological disorder caused by de novo variants in FAR1 resulting in uncontrolled synthesis of ether lipids
Quantitative Proteomics and Differential Protein Abundance Analysis after the Depletion of PEX3 from Human Cells Identifies Additional Aspects of Protein Targeting to the ER
Chornyi S, Ofman R, Koster J, Waterham H
Journal of Lipid Research 2023;64(5):100364
Journal of Lipid Research 2023;64(5):100364
The peroxisomal enzyme, FAR1, is induced during ER stress in an ATF6-dependent manner in cardiac myocytes
Marsh K, Arrieta A, Thuerauf D, Blackwood E, MacDonnell L, Glembotski C
American Journal of Physiology-Heart and Circulatory Physiology 2021;320(5):H1813-H1821
American Journal of Physiology-Heart and Circulatory Physiology 2021;320(5):H1813-H1821
An autosomal dominant neurological disorder caused by de novo variants in FAR1 resulting in uncontrolled synthesis of ether lipids
Ferdinandusse S, McWalter K, te Brinke H, IJlst L, Mooijer P, Ruiter J, van Lint A, Pras-Raves M, Wever E, Millan F, Guillen Sacoto M, Begtrup A, Tarnopolsky M, Brady L, Ladda R, Sell S, Nowak C, Douglas J, Tian C, Ulm E, Perlman S, Drack A, Chong K, Martin N, Brault J, Brokamp E, Toro C, Gahl W, Macnamara E, Wolfe L, Alejandro M, Azamian M, Bacino C, Balasubramanyam A, Burrage L, Chao H, Clark G, Craigen W, Dai H, Dhar S, Emrick L, Goldman A, Hanchard N, Jamal F, Karaviti L, Lalani S, Lee B, Lewis R, Marom R, Moretti P, Murdock D, Nicholas S, Orengo J, Posey J, Potocki L, Rosenfeld J, Samson S, Scott D, Tran A, Vogel T, Wangler M, Yamamoto S, Eng C, Liu P, Ward P, Behrens E, Deardorff M, Falk M, Hassey K, Sullivan K, Vanderver A, Goldstein D, Cope H, McConkie-Rosell A, Schoch K, Shashi V, Smith E, Spillmann R, Sullivan J, Tan Q, Walley N, Agrawal P, Beggs A, Berry G, Briere L, Cobban L, Coggins M, Cooper C, Fieg E, High F, Holm I, Korrick S, Krier J, Lincoln S, Loscalzo J, Maas R, MacRae C, Pallais J, Rao D, Rodan L, Silverman E, Stoler J, Sweetser D, Walker M, Walsh C, Esteves C, Kelley E, Kohane I, LeBlanc K, McCray A, Nagy A, Dasari S, Lanpher B, Lanza I, Morava E, Oglesbee D, Bademci G, Barbouth D, Bivona S, Carrasquillo O, Chang T, Forghani I, Grajewski A, Isasi R, Lam B, Levitt R, Liu X, McCauley J, Sacco R, Saporta M, Schaechter J, Tekin M, Telischi F, Thorson W, Zuchner S, Colley H, Dayal J, Eckstein D, Findley L, Krasnewich D, Mamounas L, Manolio T, Mulvihill J, LaMoure G, Goldrich M, Urv T, Doss A, Acosta M, Bonnenmann C, D’Souza P, Draper D, Ferreira C, Godfrey R, Groden C, Macnamara E, Maduro V, Markello T, Nath A, Novacic D, Pusey B, Toro C, Wahl C, Baker E, Burke E, Adams D, Gahl W, Malicdan M, Tifft C, Wolfe L, Yang J, Power B, Gochuico B, Huryn L, Latham L, Davis J, Mosbrook-Davis D, Rossignol F, Solomon B, MacDowall J, Thurm A, Zein W, Yousef M, Adam M, Amendola L, Bamshad M, Beck A, Bennett J, Berg-Rood B, Blue E, Boyd B, Byers P, Chanprasert S, Cunningham M, Dipple K, Doherty D, Earl D, Glass I, Golden-Grant K, Hahn S, Hing A, Hisama F, Horike-Pyne M, Jarvik G, Jarvik J, Jayadev S, Lam C, Maravilla K, Mefford H, Merritt J, Mirzaa G, Nickerson D, Raskind W, Rosenwasser N, Scott C, Sun A, Sybert V, Wallace S, Wener M, Wenger T, Ashley E, Bejerano G, Bernstein J, Bonner D, Coakley T, Fernandez L, Fisher P, Fresard L, Hom J, Huang Y, Kohler J, Kravets E, Majcherska M, Martin B, Marwaha S, McCormack C, Raja A, Reuter C, Ruzhnikov M, Sampson J, Smith K, Sutton S, Tabor H, Tucker B, Wheeler M, Zastrow D, Zhao C, Byrd W, Crouse A, Might M, Nakano-Okuno M, Whitlock J, Brown G, Butte M, Dell’Angelica E, Dorrani N, Douine E, Fogel B, Gutierrez I, Huang A, Krakow D, Lee H, Loo S, Mak B, Martin M, Martínez-Agosto J, McGee E, Nelson S, Nieves-Rodriguez S, Palmer C, Papp J, Parker N, Renteria G, Signer R, Sinsheimer J, Wan J, Wang L, Perry K, Woods J, Alvey J, Andrews A, Bale J, Bohnsack J, Botto L, Carey J, Pace L, Longo N, Marth G, Moretti P, Quinlan A, Velinder M, Viskochil D, Bayrak-Toydemir P, Mao R, Westerfield M, Bican A, Brokamp E, Duncan L, Hamid R, Kennedy J, Kozuira M, Newman J, Phillips J, Rives L, Robertson A, Solem E, Cogan J, Cole F, Hayes N, Kiley D, Sisco K, Wambach J, Wegner D, Baldridge D, Pak S, Schedl T, Shin J, Solnica-Krezel L, Waisfisz Q, Zwijnenburg P, Ziegler A, Barth M, Smith R, Ellingwood S, Gaebler-Spira D, Bakhtiari S, Kruer M, van Kampen A, Wanders R, Waterham H, Cassiman D, Vaz F
Genetics in Medicine 2021;23(4):740-750
Genetics in Medicine 2021;23(4):740-750
Quantitative Proteomics and Differential Protein Abundance Analysis after the Depletion of PEX3 from Human Cells Identifies Additional Aspects of Protein Targeting to the ER
Zimmermann R, Lang S, Lerner M, Förster F, Nguyen D, Helms V, Schrul B
International Journal of Molecular Sciences 2021;22(23):13028
International Journal of Molecular Sciences 2021;22(23):13028
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
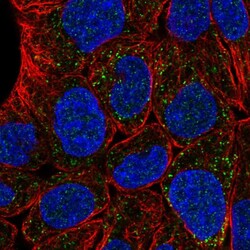
- Experimental details
- Immunofluorescent staining of human cell line RT4 shows localization to peroxisomes.
- Sample type
- Human
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry